The “Knowledge Networks in Biomedical and Behavioral Research” meeting organized under the auspices of the NIH Knowledge Graph Working Group was held at the NIH Neuroscience Center (NSC) building in North Bethesda, Maryland on May 28-29, 2025. The meeting brought together thought leaders and leading scientists in this field, who are PIs or key personnel of NIH-funded projects, NIH program officers and program officials of the NSF partnership program “Proto-Open Knowledge Network (Proto-OKN)”. The meeting was co-chaired by Cathy Wu (U. Delaware) and Chunlei Wu (Scripps Institute). There was a total of 88 registrants including NIH program staff.
Knowledge Graphs (KG) have become a critical tool for knowledge discovery in a computable and explainable manner. Due to the semantic and structural complexities of biomedical data, biomedical knowledge graphs need to enable dynamic reasoning over large evolving graphs and support fit-for-purpose abstraction through establishing standards, preserving provenance and enforcing policy constraints for actionable biomedical discovery. To explore the opportunities, challenges, and future directions of a biomedical knowledge network, the main goals of the meeting were:
- to identify opportunities and challenges in KG research;
- to discuss strategies to facilitate developments in artificial intelligence (AI) technologies and approaches for building KGs; and
- to develop guidelines for ethical, transparent, and informed use/reuse of data, knowledge, and models.
Meeting discussions led to defining the following six desired/required properties of a biomedical knowledge network:
- inference and reasoning in biomedical knowledge network need domain-centric approaches;
- harmonized and accessible standards are required for knowledge network representation and metadata;
- robust validation of biomedical knowledge graphs needs multi-layered, context-aware approaches that are both rigorous and scalable;
- the evolving and synergistic relationship between knowledge graphs and large language models is essential in empowering AI-driven biomedical discovery;
- integrated development environments, public repositories, and governance frameworks are essential for secure and reproducible knowledge network sharing; and
- robust validation, provenance, and ethical governance are critical for trustworthy biomedical knowledge networks.
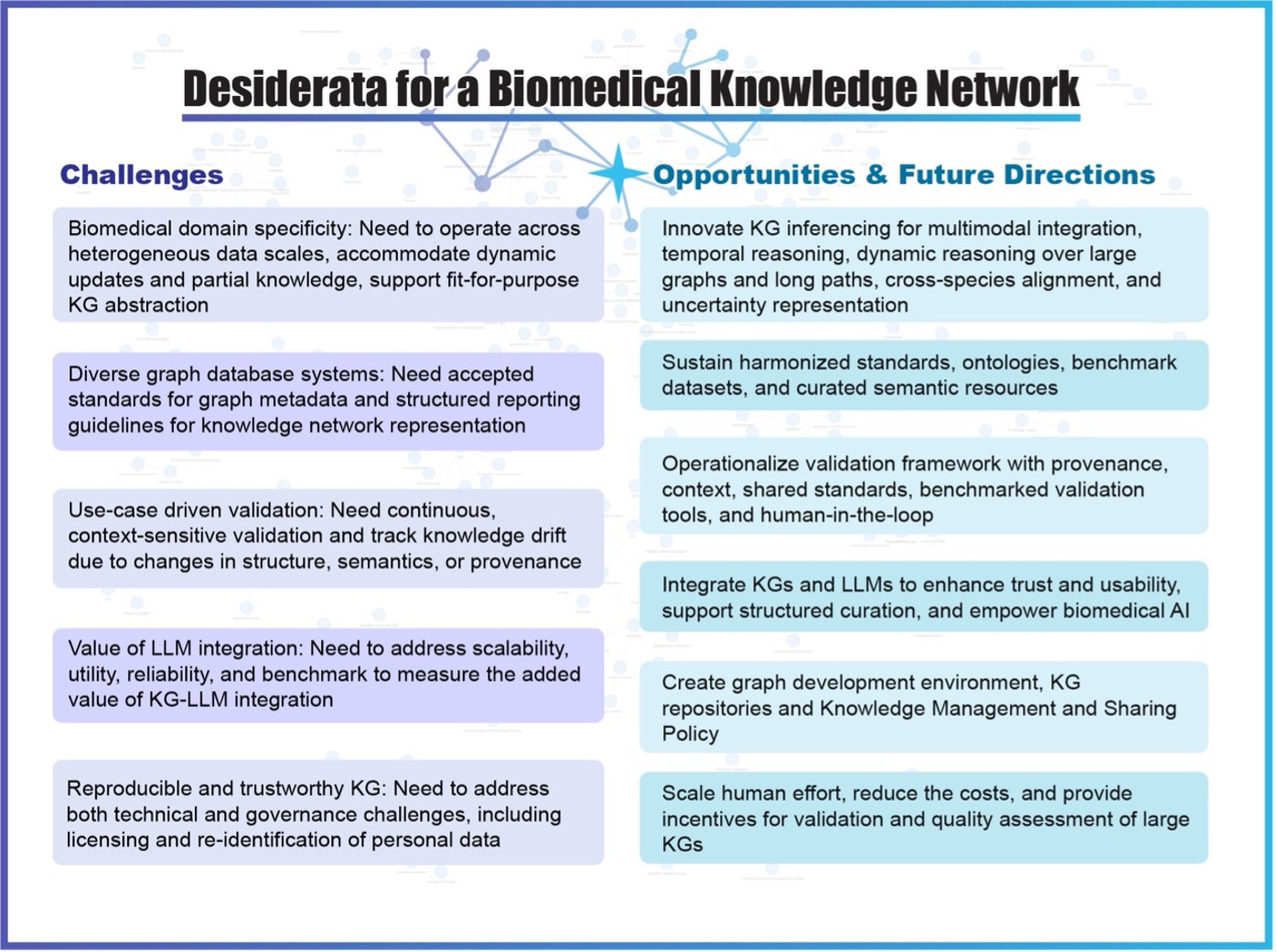
Realizing the full potential of a biomedical knowledge network requires a principled fusion of domain specificity, semantic rigor, computational scalability, reproducibility and trustworthiness. Meeting participants identified several challenges as well as opportunities for building a (NIH- or even larger-scale) biomedical knowledge network that is capable of supporting robust, interpretable, and actionable biomedical discovery. It was also noted by the participants that the virtues of the biomedical knowledge network lie in its trustworthiness. The concept of ‘trustworthy’ can be described as epistemic components, relating to provenance and validation, and ethical components, such as privacy and security protections. It was also emphasized that as knowledge graphs become more central to biomedical AI, ensuring their integrity and social acceptability will be as important as ensuring their computational power.
Figure below summarizes the Challenges and Opportunities in biomedical knowledge network research:
Meeting Organization
Co-chairs: Cathy Wu and Chunlei Wu
Session co-leads: Hongfang Liu, Jason Flannick, Mark Musen, Andrew Su, Lawrence Hunter, Thomas M Powers
Session panelists: Chris Bizon, Katy Börner, Licong Cui, Nick Evans, Jeffrey Grethe, Melissa Haendel, Bian Jiang, Chris Mungall, Jonathan Silverstein, Deanne Tayler, Chunhua Weng


