Tuesday, July 1, 2025

By: Dr. Susan Gregurick, Associate Director of Data Science, NIH, and Hsinyi (Steve) Tsang, PhD, Clinical Informatics Lead, Office of Data Science Strategy, NIH
Imagine you are doing a longitudinal study on food allergy prevalence and severity in children. You want to understand if there have been any noticeable changes in allergy patterns and reactions going back 5, 10, and 15 years. As a researcher working with the National Institutes of Health (NIH), you have access to a lot of data about food allergies—which is great because you need a lot of data for your study.
But there is a problem: The data comes from NIH studies and different institutions nationwide. And, the researchers conducting these earlier studies used different methods to document and classify allergic reactions, making what should have been a relatively straightforward task complicated. Now, you need to figure out how to harmonize data where some studies used numerical severity scales, others used descriptive categories (i.e., mild/moderate/severe), and still others focused on specific clinical manifestations like IgE levels and skin test wheal size. Unfortunately, this is a very real headache for many biomedical researchers. Fortunately, it's a headache that NIH is trying to fix, and we're using generative artificial intelligence (GenAI) to help us do it!
At the core of this challenge is a lack of Common Data Elements (CDEs). CDEs are building blocks for research studies that ensure biomedical data is FAIR (Findable, Accessible, Interoperable, and Reusable). In practice, this means researchers ask the same question about allergy symptoms, generating the same type of data, or ensuring saved data can be accessed by different types of computer operating systems.
Promoting wider usage of CDEs is foundational to the work of NIH’s Office of Data Science Strategy (ODSS) and plays a central role in the final 2025-2030 NIH Strategic Plan for Data Science. The strategic plan highlights the need to create minimal sets of consistent and computable CDEs that support data integration across studies and repositories. It calls out programs that are already championing CDEs, such as the Mobile At-Home Reporting through Standards (MARS) and the Helping to End Addiction Long-Term (HEAL) initiative. It also emphasizes ongoing efforts to expand the usage of CDEs, including the NIH CDE Repository, NCI’s Enterprise Vocabulary Services, and the Cancer Data Standards Registry and Repository (caDSR). The strategic plan reaffirms NIH’s commitment to making CDEs a core aspect of our research enterprise.
NIH is not alone in pushing for broader adoption of CDEs. Congress appropriated funds in Fiscal Year 2024 that directed NIH and ODSS to “encourage development and use of CDEs in disease areas where they currently do not exist.” In response, ODSS did a public workshop and sent out a request for information to engage the research community on the matter of CDEs. ODSS also co-funded projects with NIH Institutes and Centers on a variety of diseases and disease areas, including chronic diseases, childhood diseases, and neurological diseases.
In parallel, ODSS sought to understand the extent of NIH’s existing efforts to promote the adoption and usage of CDEs across the entire research ecosystem. Conducting a landscape analysis across such a vast biomedical field is nearly impossible, but we leveraged the power of GenAI to help with the task.
For this landscape analysis, ODSS used GenAI to rapidly synthesize large amounts of data, taking a vast landscape and making it bite-sized for easier analysis. This is partly possible because GenAI’s predictive modeling capabilities helped streamline the analysis by anticipating patterns in publications. This approach enabled the ODSS team to categorize and identify areas of focus for approximately 145 publications that leveraged CDEs—dating back to 1978.
For this landscape analysis, the ODSS team utilized the National Institute of Allergy and Infectious Diseases’ (NIAID) government-approved GenAI Platform, DocBot. It is built on Microsoft’s implementation of OpenAI’s AI model, hosted within NIH’s Azure environment, and accessible through STRIDES. DocBot excels at reading documents and providing answers through conversational chats with users. DocBot reviewed publications and notices of funding opportunities (NOFOs) for this analysis, while the human team members reviewed awards. The team also supported the AI by providing frameworks and occasional interventions to ensure accurate data analysis. And finally, the team ensured all the data processed by the GenAI was publicly available, open access, and free of any Personally Identifiable Information (PII) or Protected Health Information (PHI). This approach allowed the team to look at the landscape from multiple perspectives, including by scientific topic, funding institute, and even fiscal year.
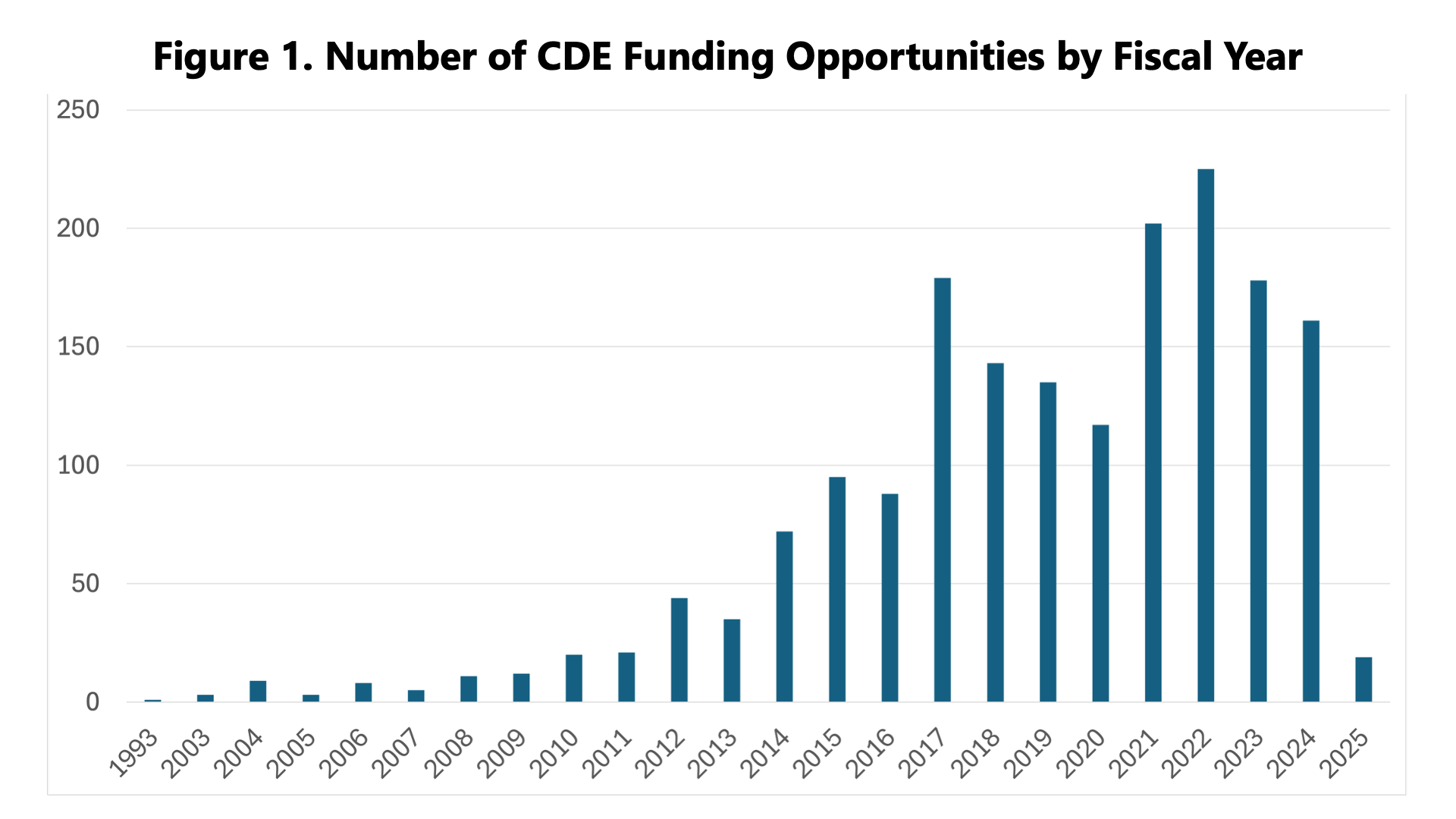
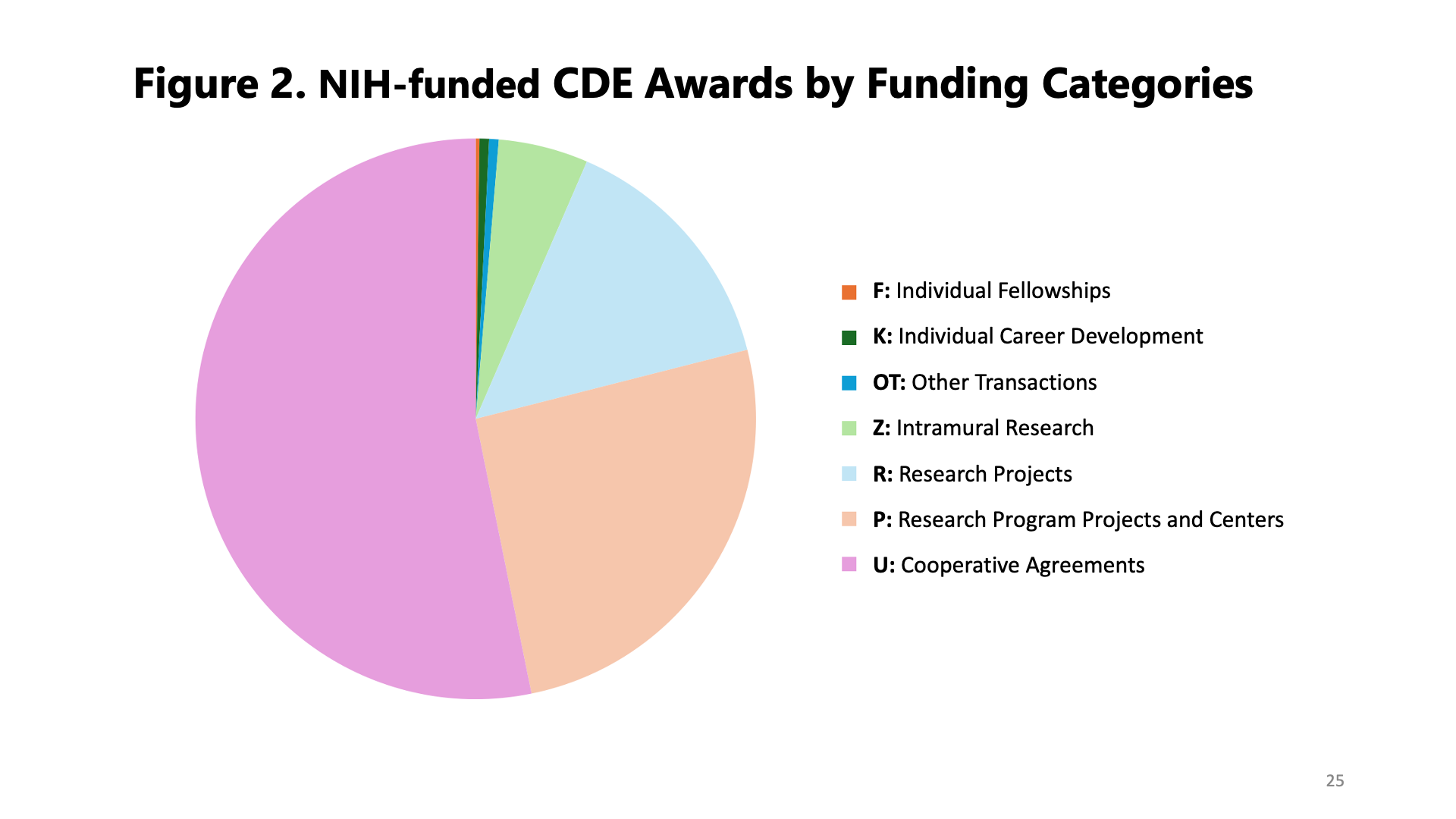
The preliminary findings revealed a fuller picture of CDE usage at NIH, calling out interesting trends in our work with CDEs to date and potential areas for further investment in the future (and with alignment to the NIH Strategic Plan for Data Science). So far, funding and awards for projects that use CDEs are cyclical, with a marked increase in the last 10 years (Figure 1). Additionally, most of the funding has been awarded to core facilities or data coordinating centers, meaning research projects where the data must be shared among NIH-supported researchers (Figure 2).
Finally, regarding publications, the GenAI analysis validated our hypothesis that most of the CDE publications primarily focused on the application of CDEs in clinical and translational research. The next most prominent areas included publications on developing and vetting CDEs—which highlighted collaborative and iterative processes involving expert review and public feedback—and standardization and harmonization—which focused on aligning CDEs across domains to support consistent data collection and integration of datasets.
Although the current body of literature offers a strong foundation demonstrating the value of CDEs, including their applications in AI and machine learning, this remains a rapidly evolving field. Some other emerging areas of focus are the use of CDEs in community research and strategies for reusing existing CDEs. Ongoing research is needed to address emerging challenges and to broaden the adoption and impact of CDEs across diverse research settings.
To that last point, the ODSS team continues to refine our GenAI analysis. In response to our findings, we have identified key lessons organized into a “Three C’s” framework to promote the adoption and use of CDEs.
First, foster more collaborations between Institutes and Centers within NIH that prompt researchers to use CDEs for better data sharing. Second, establish a community of practice around CDE usage that helps pool insights and build research capacity. Third, communicate about CDEs to raise awareness and provide strategies that promote CDE adoption.
Additionally, the ODSS team recognized the potential GenAI tools hold in achieving more with fewer resources, enabling teams to engage in smarter, more efficient meta-analysis of large data sets. This analysis marked an important first step in harnessing AI’s full potential for data science at NIH. Now, we are looking at how we can leverage GenAI with other workflows, improving the scale and reusability of this emerging tool.
To learn more about CDEs you can visit the CDEs webpage on the ODSS website. You can learn more about NIH’s AI strategy by visiting the Office of Science Policy website.


